A New Methodology for Protein Design
A New Methodology for Protein Design
MIT Department of Biology, Whitehead Institute, MIT Department of Biological Engineering, Koch Institute at MIT
The structure of a protein is a product of its amino acid sequence and environment. While in theory the number of potential configurations available to a protein is greater than the number of atoms in the observable universe, in practice it has been observed that most proteins adopt strikingly similar folds. Recently, it has been shown that the structures of all known proteins can be decomposed into a set of frequently occurring backbone tertiary motifs, named “TERMs”, which can be combined to produce the backbone structure of any given protein.
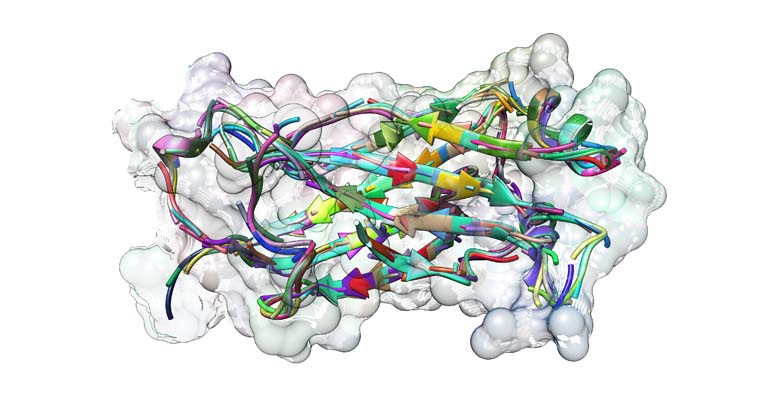
This image captures a superposition of the smaller structural motifs that make up the complete structure of a common protein, TNF-associated factor 6 (TRAF6). TRAF6 is a signaling protein with a critical role in inflammatory responses. Inhibition of TRAF6 has demonstrated potential for treating inflammation associated with illnesses such as multiple sclerosis and cardiovascular disease.
By decomposing a protein structure into smaller motifs, we gain access to a wealth of existing information accumulated over the years. Through this new lens, each protein is viewed not as a wholly new instance of structure, but as an arrangement woven from compatible building blocks, drawn from a finite set. By generating TERM-based representations of proteins, we can learn the algorithm by which nature arranges these motifs in natural proteins and harness them for design.